Research
Our research lab focuses on developing theoretical and computational methods to study biological processes across different temporal and spatial scales. We integrate physics, multiscale simulations, and machine learning to unravel the mechanisms of essential biological machineries and design molecules for treating related diseases. We strive to build a diverse team, bringing together a wide range of backgrounds and expertise.
Multiscale modeling and simulations of biological machineries
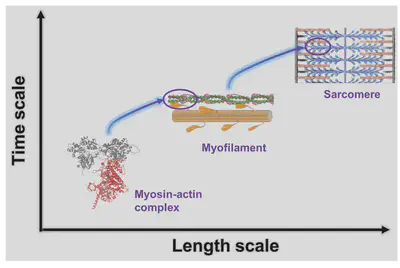
Our lab focuses on developing and applying multiscale modeling methods to connect molecular biology with systems biology, harnessing the power of modern computational physics. For example, we are building a multiscale model that characterizes how small changes in contractile proteins impact cardiac sarcomere function, based on our previous work (PNAS 2023, PNAS Nexus 2024)
Applying deep learning and statistical physics to reveal protein conformations and functions
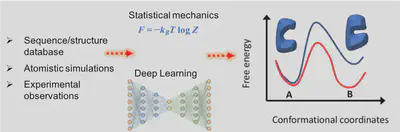
Molecular simulations have become a valuable complement to laboratory and clinical studies in combating various diseases. We are interested in applying statistical physics and deep learning to extend the limited timescales that conventional molecular simulations are able to cover. Previously, we employed rare-event sampling techniques to search for the most probable transition pathways for molecular motor actions (JACS 2015, eLife 2018).
Drug discovery inspired by multiscale simulations
Taking advantage of the conformational ensembles obtained from the aforementioned simulations, we discover potential drug binding sites and screen allosteric modulators with the assistance of machine learning. We are currently designing small molecules targeting cardiac myosin (for cardiomyopathy) and LRRK2 (for Parkinson's disease, ACS Chem Biol 2023, Cell Discovery 2024).